Cytokines play essential signaling functions in the immune system, enabling both short and long-range communications between tissues and the immune system, as well as between immune cells.
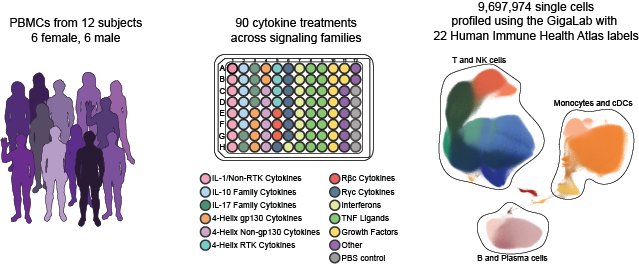
Parse Biosciences utilized their GigaLab platform to profile 90 cytokine perturbations in peripheral blood mononuclear cells (PBMCs) from 12 human donors. After sequencing, data assembly, and quality control with the Parse Analysis Pipeline, the resulting dataset consists of transcriptomic profiles from ~10 million PBMCs.
See the Parse Biosciences dataset page to get more information about how this dataset was generated:
For more in-depth analyses, see the manuscript by Oesinghaus, Becker, Vornholz, and Papalexi, et al.:
Note: these authors are not affiliated with the Allen institute.
At the Allen Institute, we frequently observe the effects of cytokine signaling in our human cohort data, and cytokines are known to play key roles in both immune health and disease.
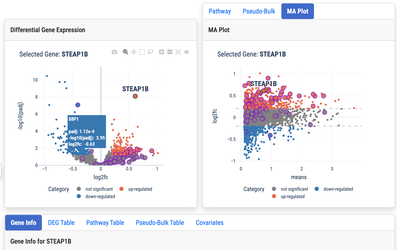
To better understand the effects of each cytokine on PBMCs, we downloaded the dataset from Parse Biosciences, labeled the data using the cell types in our Human Immune Health Atlas, then performed differential expression tests to identify genes that respond to each cytokine. The CellTypist labeling models, DEG results, and gene sets suitable for use in gene set enrichment analysis (GSEA) are available from the Downloads page.
To enable exploration of this dataset, we applied our UMAP and DEG visualization tools to the results of our analysis:
Visualize differential expression and gene set enrichment for comparisons between all 90 cytokines and PBS controls across 22 PBMC cell types.
Explore Differential ExpressionExplore the dataset downsampled to 5M cells in a UMAP projection.
Explore UMAP Visualization