Configure a Pipeline (Tutorial)
 | Abbreviations Key |
| CBC | complete blood count |
| CSV | comma-separated values |
| EMR | electronic medical record |
| HISE | Human Immune System Explorer |
| IDE | integrated development environment |
| QC | quality control |
| REDCap | Research Electronic Data Capture |
At a Glance
This document outlines how to ingest lab results, surveys, and patient history into HISE. It describes the pipeline for data ingestion, project configuration requirements, and options for simple and complex configuration. The document also covers data curation features, including pipeline approvals and duplicate batch identification, available to select users.
 Ingest Lab Results
Ingest Lab Results
Lab results you submit to HISE are ingested as CSV files and become part of the sample metadata. The ingest pipeline checks for missing and illegal values, normalizes the results to adhere to common names and value range standards, and associates the results with the correct samples. Follow the instructions below to work with your ingested lab results.
Instructions
1. Navigate to HISE, and use your organizational email address to sign in.
2. Configure your project. The two available configuration options, simple and complex, are described in the following sections. In both cases, you can submit partial data sets in separate ingests, to add CBC data incrementally. It is also possible to cancel out or overwrite data (see Step 2C).
A. Simple configuration. Contact immunology-support@alleninstitute.org to define your expected data before ingestion. In most cases, the definition is straightforward and includes the following information:
i. Associated patient and sample data
ii. CSV column header in which the data is to be ingested
iii. Friendly name to be used in HISE
iv. Preferred units of measure
v. Normal range of values for ingested data
B. Complex configuration. If naming conventions or ranges need to be standardized and configuration is more complex, more extensive project setup might be necessary.
C. (Optional) Resubmission. To retry a lab results ingest after fixing an error, ingest the updated lab results CSV file. To overwrite previously ingested CBC data, make sure the file name includes retry, Retry, RETRY, or ReTrY.
3. View your ingested results.
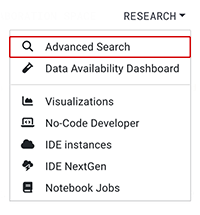
A. From the top navigation menu, choose RESEARCH > Advanced Search.
B. On the Query Builder page, click one of the listed parameters, such as SAMPLE METADATA.
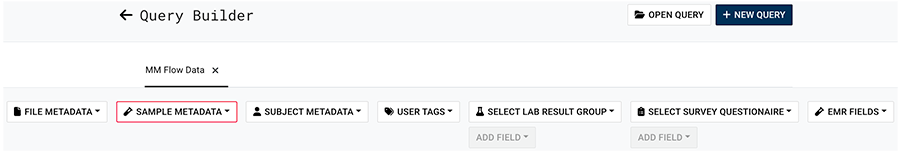
C. From the drop-down list, choose a field, such as PANEL ID.
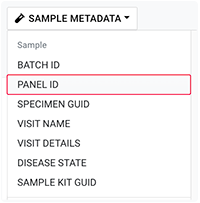
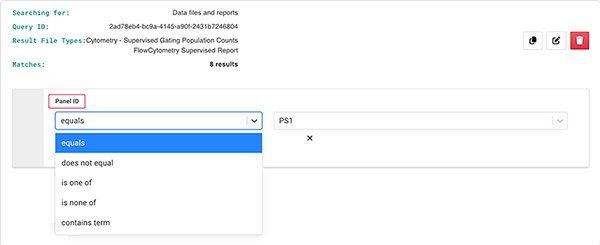
D. In the dialog box, add a search operator for the selected field (in this case, Panel ID).
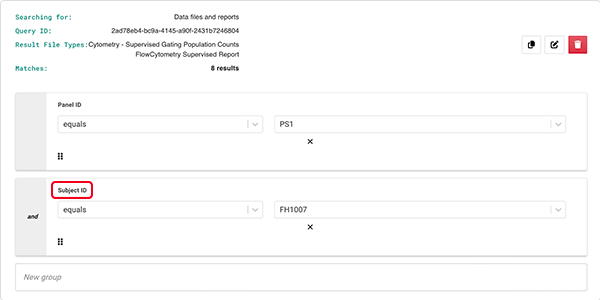
E. (Optional) To narrow your search, choose additional parameters and fields (in this case, Subject ID).
F. On the Search Results page, in the far left column, select one or more checkboxes corresponding to the results you want to see. Alternatively, to see all results, select the checkbox at the top of the column.
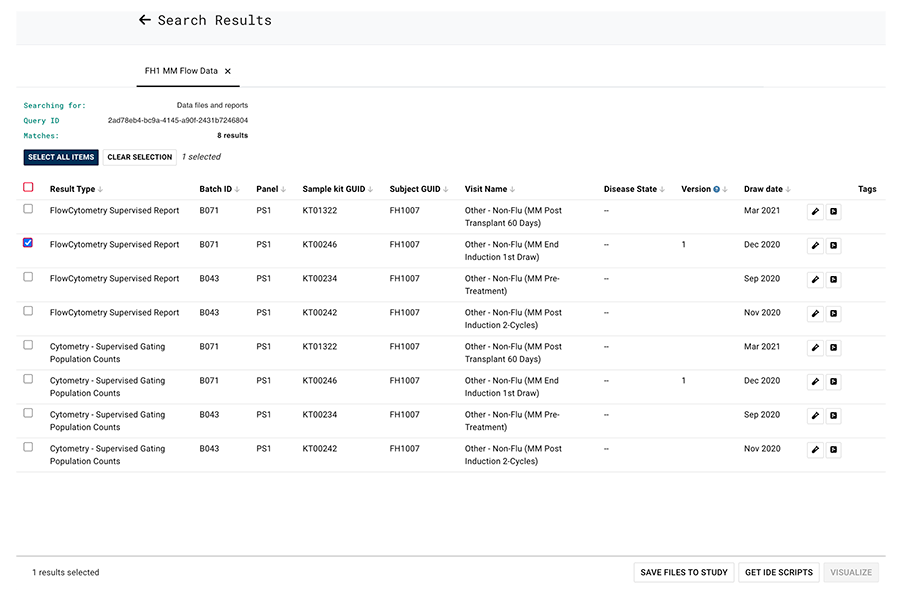
G. In the bottom-right corner of the Search Results page, click an available option. (Some options might not be available. For example, the VISUALIZE option is grayed out in the current query.)

i. To save the results and associate them with a study, choose SAVE FILES TO STUDY.
ii. To read the results into an IDE, choose GET IDE SCRIPTS.
iii. To visualize the data, choose VISUALIZE.
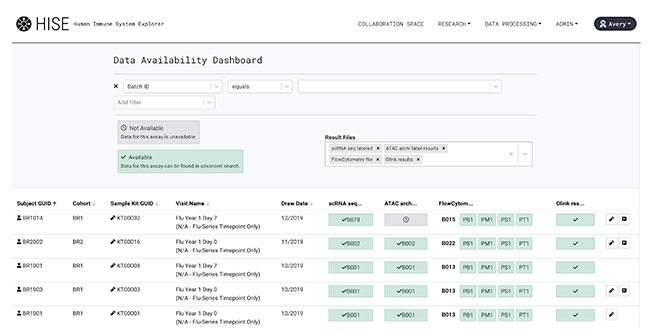
4. (Optional) View your results in the Data Availability Dashboard.
 Ingest Patient History Data
Ingest Patient History Data
You can use CSV files to ingest survey, questionnaire, or EMR data into HISE.
Ingest Survey or Questionnaire Data
Survey or questionnaire data is submitted to HISE in CSV files, validated, and stored as sample metadata. The ingest pipeline aligns the data with the original questionnaire and associates the results with the correct samples. As with ingested lab results, survey/questionnaire data is displayed in the Data Availability Dashboard, or you can use it to select data in an advanced search. Survey results can also be read into the IDE, either embedded in a results file or ingested separately as file descriptors only.
Your survey or questionnaire design must be declared in the project before you ingest data. Contact immunology-support@alleninstitute.org for help using the REDCap data dictionary to define the data for export and ingestion into a watchfolder.
Ingest EMR Data
Patient history (EMR) data is submitted to HISE in CSV files, validated, and stored as subject metadata. The ingest pipeline associates the results with the correct subject and hospital visit. The EMR data is displayed in the Data Availability Dashboard and can be used to select data in an advanced search. The metadata can also be read in the IDE.
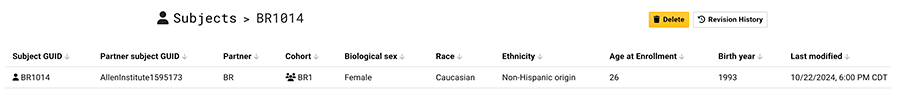
A subject's patient history may span multiple hospital visits. Each separate visit is denoted with a visit date, which can include either the year or the month and year. In addition, each separate visit must contain a "days since first research visit" value, expressed as a number of days. For instance, if the first hospital visit was 7 days before the first research visit, the "days since first research visit" value is 7. If multiple hospital visits are recorded in the same year and month, the "days since first research visit" distinguishes one visit from another.
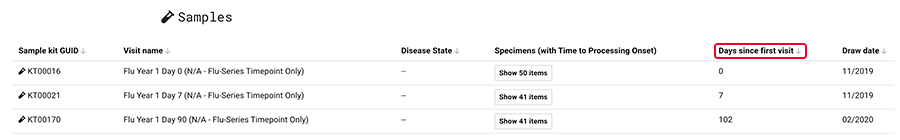
Your patient history data must be declared in the project before you ingest data. Contact immunology-support@alleninstitute.org for help with project setup.
 Curate Pipeline Data
Curate Pipeline Data
Select users have access to pipeline data curation options.
Pipeline approvals
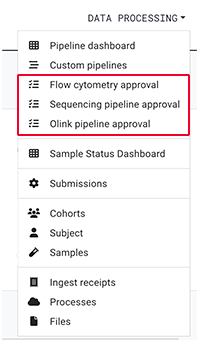
To approve assay pipelines results before they're released to the RESEARCH space, click DATA PROCESSING > [Selected pipeline]. There are specialized views for flow cytometry, sequencing, and Olink.
Duplicate batch identification
When two pipelines are run on the same raw data, either because a prior run was rejected or because an error occurred, the results of the older run are automatically hidden. However, when two separate runs are conceptual duplicates but actually ingest different raw data sets, HISE doesn't automatically recognize the runs as potential duplicates, and both sets of results appear. For instance, to correct a pre-ingestion irregularity or to resequence data at a new preferred depth, the wet lab might create a second raw data set using the same underlying material. In such cases, the same approval views can be used to mark the runs at duplicates, thereby removing the older results, as well as QC reports and other pipeline deliverables, from the RESEARCH space. To request that a run be marked as a duplicate, contact immunology-support@alleninstitute.org.
 Related Resources
Related Resources
View the Sample Status Dashboard (Tutorial)