Last modified 2025-10-06 |
Explore IDE Folders
 | Abbreviations Key |
| HISE | Human Immune System Explorer |
| HTML | hypertext markup language |
| IDE | integrated development environment |
| SDK | software development kit |
| UI | user interface |
At a Glance
This document explains the HISE IDE workflow, including the function and persistence of each folder in your IDE. When you're ready to get started, see Create Your First HISE IDE (Tutorial). To learn how to start, stop, restart, delete, or clone an IDE instance, see Use HISE IDEs (Tutorial). If you have questions, contact Support .
IDE Basics
An IDE is a comprehensive software suite that consolidates various scientific tools and libraries into a unified platform. Working in a HISE IDE streamlines the data analysis process by providing a centralized space in which to work and visualize your data. You can choose from preinstalled data modalities curated by experienced software developers with the help of leading scientists in flow cytometry, Olink protein analysis, scRNA-seq, single-cell genomics, and other specialized areas. For details about data modalities, see Manage IDE Packages.
Folder Hierarchy
The folders in a HISE IDE (left) are described below and summarized in the image (right). The table below summarizes folder persistence.
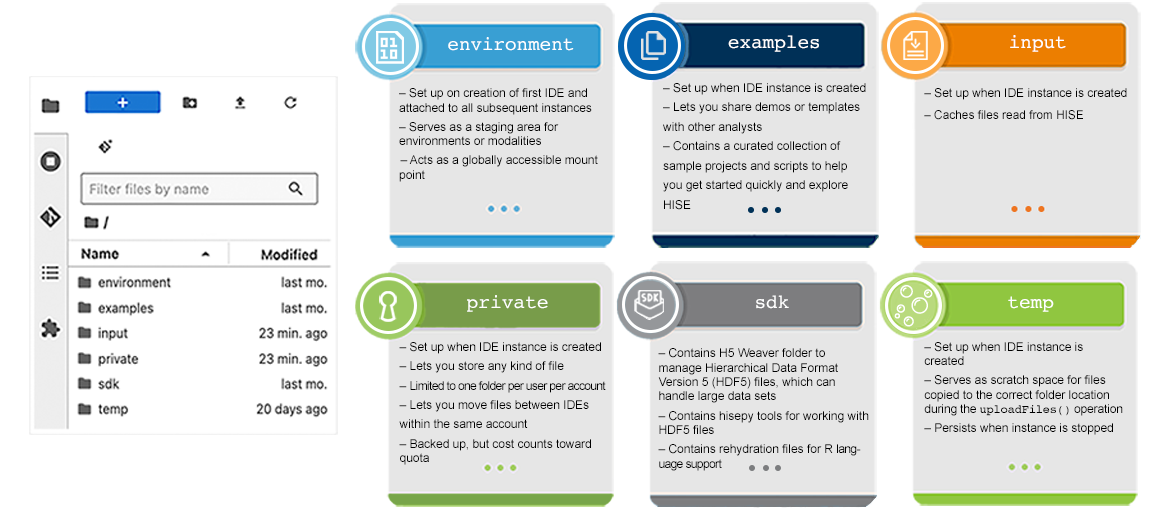
Environment
The /environment folder contains the Conda environment for the modality you select, as well as all necessary dependencies. If you use conda install <package name>, the package is added to the /environment folder.
Examples
The /examples folder is a space for analysts and developers to share demos and templates—primarily in HTML or Markdown format—with other analysts. To make an example available in the IDE, add it to the /demos folder. Any file added to that folder is automatically copied to the /home/jupyter/examples IDE folder on startup. Files outside the /demos folder are not copied.
| Take great care not to include real data in your examples. To safeguard protected health information, data must stay within the account in which it originated. Think of an account as a walled garden and the data as plants that must never cross-pollinate with plants from surrounding walled gardens. (In this metaphor, bees do not exist.) |
Thanks to this collaborative process, the /examples directory now offers a curated collection of sample projects and scripts that demonstrate how to use HISE SDK methods in R or Python. The folder also contains ready-to-run sample Dash applications and other examples to help you visualize data in HISE.
Input
The read-only /input folder is used to store incoming data and system configuration files needed for proper execution. A caching mechanism allows temporary storage of files to speed up loading and improve performance when frequently accessed data is retrieved.
| Folder | Persists? (Y/N) |
/environment | Y |
/examples | Y |
/input | N |
/private | Y |
/sdk | N |
/temp | Y |
Private
When it doesn’t make sense to write your outputs or intermediates into HISE, the /private folder stores your data. This folder can also be used to move content between IDEs within the same account. Files written into the /private folder folder are placed into cheap, durable cloud storage that persists even after the IDE is deleted.
SDK
The SDK is a collection of tools for expanding the HISE platform by adding visualizations, uploading new findings, or performing other specialized tasks. The SDK also supplies methods that let you interact with HISE programmatically in either R or Python. These methods are available once your VM is created, so there’s no need to install additional tools. For details, see Use HISE SDK Methods. The tools stored in the /sdk folder do not persist when the instance is stopped.
Temp
The /temp folder is used to cache data in active use, process temporary files, and store intermediate computational results. This data persists when the instance stops.
 Related Resources
Related Resources