Last modified 2025-10-23 |
Use the Read Files SDK Method (Tutorial)
 | Abbreviations Key | |||||
bool | boolean | IDE | integrated development environment | |||
df | DataFrame | SDK | software development kit | |||
desc | descriptor(s) | pd | pandas | |||
dict | dictionary | tmp | temporary | |||
| HISE | Human Immune System Explorer | UUID | universally unique ID | |||
hp | hisepy | |||||
At a Glance
This document explains how to use read_files() to download files to your HISE IDE. If you have questions, contact Support.
Method signature
 read_files()
read_files()
hp.read_files(
file_list: list = None,
query_id: list = None,
query_dict: dict = None,
to_df: bool = True,
)
 readFiles()
readFiles()
readFiles( fileIds = list | NULL, queryId = character(1) | NULL, query = list | NULL ) → list<data.frame> </data.frame>
Parameters
The parameters for this method are listed in the following table. In each key:value pair, the value must be of type list.
Python Parameters | ||||
| Parameter | Data type | Description | ||
file_list | list | List of UUIDS to retrieve | ||
query_id | string | Value of the queryID from an advanced search | ||
query_dict | dict | Dictionary that allows users to submit a query | ||
to_df | bool | Boolean determining whether the result is returned as a DataFrame | ||
R Parameters | |||
| Parameter | Description | ||
fileIds | List of UUIDS to retrieve | ||
queryId | UUID from an advanced search | ||
query | List of query params to search for. The format is similar to that passed to getFileDescriptors, but the fields correspond to fields in the Subject materialized view. NOTE: fileType with a valid entry must be present | ||
Description
This function fetches HISE files and returns one or more objects when you pass in the following:
- A list of file IDs (Python:
file_list| R:fileIds) - A saved search ID (
query_id| R:queryId) - A custom search query (
query_dict| R:query)
The object returned is either a dictionary (Python) or a list of data.frames (R). The dictionary or list contains keys [descriptors, labResults, specimens, values].
Instructions
The following instructions are written for Python. To adapt them for R, use the R function signature and parameters listed above.
 Import libraries
Import libraries
To get started, set up your environment to interact with HISE programmatically and access all available SDK functions. For details, see Use Hise SDK Methods and Get Help in the IDE.
1. Navigate to HISE, and use your organizational email address to sign in.
2. Open an IDE. For instructions, see Create Your First HISE IDE (Tutorial).
3. For programmatic access to HISE functions and efficient handling of tabular data, import the Python SDK and the pandas library.
# Import hisepy and pandas
import hisepy as hp
import pandas as pd
 Define file IDs
Define file IDs
In this step, we define the file IDs for this notebook. For details, see Use Advanced Search (Tutorial).
1. Retrieve your own set of file IDs, and then define them as shown below. (The example below uses placeholder UUIDs—replace them with your own.)
# Define the file IDs used in this analysis
FILEIDS = ['4551e620-48db-4328-a2b0-122730cd128d', '6417a4c5-098b-4d70-8c24-951e1c1c44ce']
 Return dictionary output and apply tabular format
Return dictionary output and apply tabular format
| To see what's in a given dictionary key, use the following format:
|
When you call read_files() with the to_df=True parameter, a dictionary is returned in which each key contains a pandas DataFrame. The to_df=True parameter arranges the data into a tabular format for easier analysis.
1. Pass your list of file IDs toread_files().
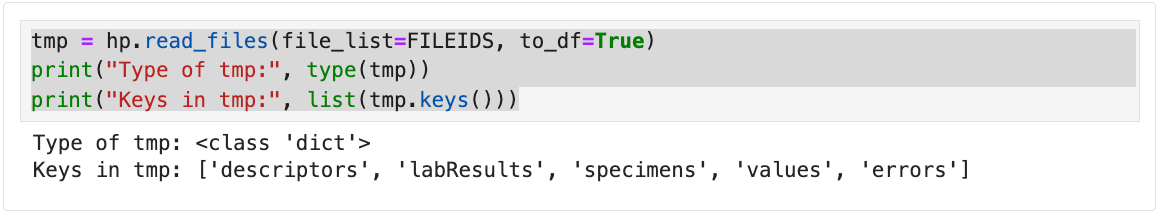
# Return dictionary output and print keys from read_files
tmp = hp.read_files(file_list=FILEIDS, to_df=True)
# Shows the class of the returned object
print("Type of tmp:", type(tmp))
# Prints all keys (file IDs or names) in the dictionary
print("Keys in tmp:", list(tmp.keys()))
The following output is returned.
 Preview the data
Preview the data
Each key in the tmp dictionary represents a different dataset returned by hp.read_files(). The accompanying table summarizes the content of each key.
| Key | Description |
|---|---|
descriptors | Project, sample, or subject metadata |
labResults | Test results and IDs |
specimens | Status and info on biological specimens |
values | Raw data metrics |
errors | File retrieval errors, if any |
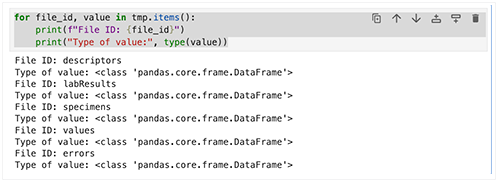
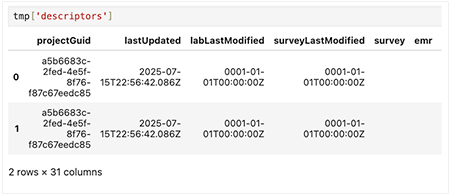
1. For each key, use a loop to print the file ID, the value type, and a preview of the data.
for file_id, value in tmp.items():
print(f"File ID: {file_id}")
print("Type of value:", type(value))
# If it's a DataFrame, show the first few rowstry:
print(value.head())
except AttributeError:
print(value) # For non-DataFrame types
print("-" * 40)
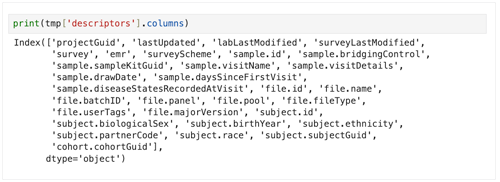
2. To see all column heads for a given data set, use the following line.
print(tmp['descriptors'].columns)
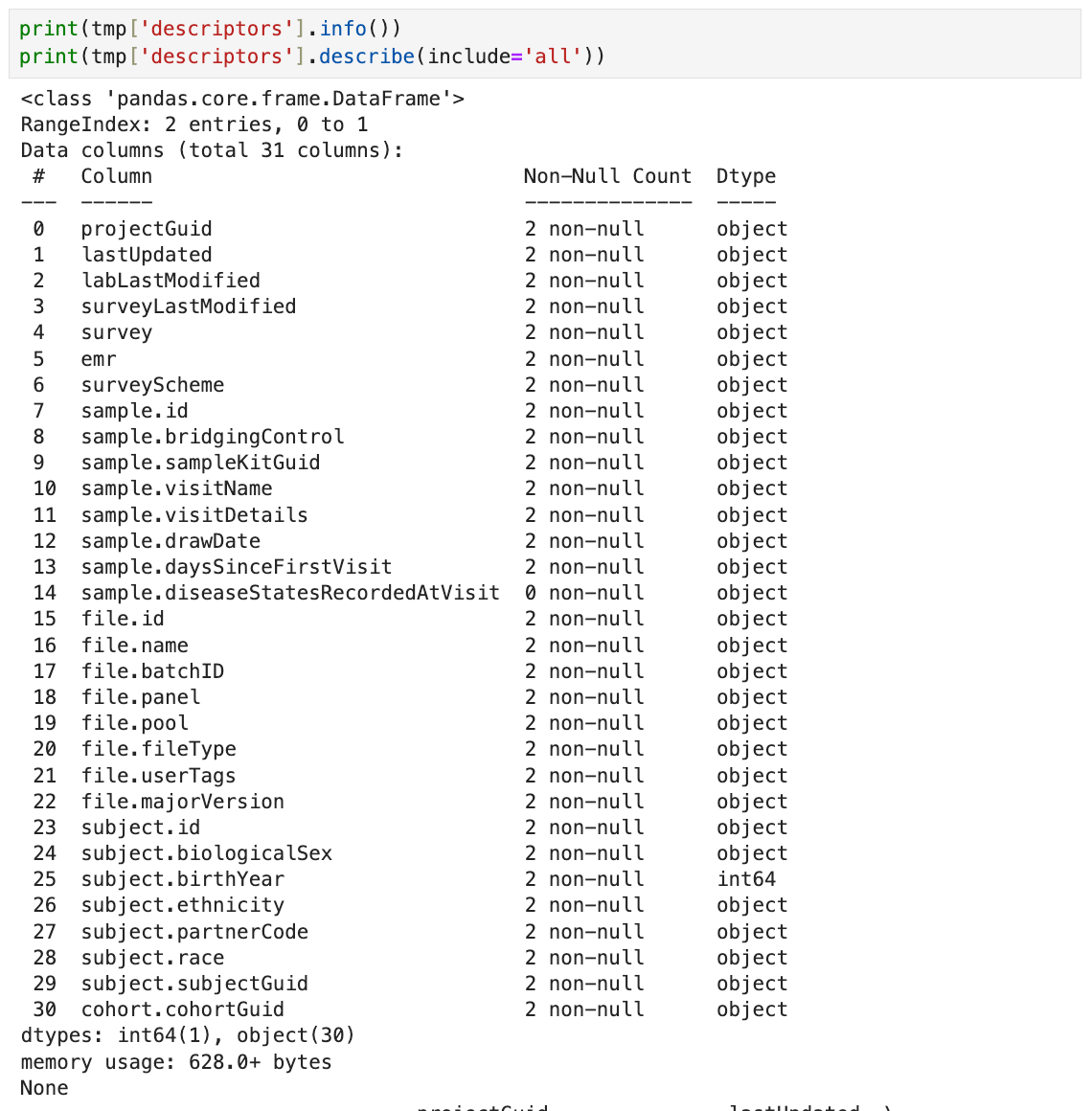
3. To get a summary of the DataFrame, use the following line.
print(tmp['descriptors'].info())
print(tmp['descriptors'].describe(include='all'))
 Related Resources
Related Resources