| Last modified 2025-04-14 |
Q&A and Troubleshooting Guide
 | Abbreviations Key |
| AIFI | Allen Institute for Immunology |
| HISE | Human Immune System Explorer |
| IDE | integrated development environment |
| UI | user interface |
| VM | virtual machine |
At a Glance
AIFI now offers IDEs designed specifically for the needs of our scientists and data analysts. These new IDEs are faster and easier to spin up and navigate. They support a workflow that promotes reproducibility, collaboration, and transparency. This document answers frequently asked questions about HISE and the NextGen IDEs. For specific issues, contact immunology-support@alleninstitute.org, or click the Support button in the upper-right corner of this document.
Jump to:
Troubleshooting
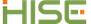
Q: I'm getting an uploadFiles error saying that the input file is not cached even though I just cached in the preceding cell.
A: This is a known issue: When you upload a result, there's a lag before the result is indexed, which makes it available in Advanced Search. Until we can eliminate that lag time, the workaround is to run cacheFiles() and then try your uploadFiles call again.
Q: My IDE shows a startup script failure. I get a Failed to stop message when I try to stop or delete the IDE. My other IDE reaches the idle timeout error after about 30 minutes even though the limit is set to 24 hours. What's the deal?
A: You've probably run out of space in your home directory. Dump a couple of gigs' worth of files in said directory, and retry your startup. You're welcome.
Q: I can’t figure out how to get my Scanpy/Matplotlib plots to show up. I tried them “vanilla,” I tried adding plt.show() and matplotlib.use(`Agg`), but nothing worked.
A: To enable inline plotting of Matplotlib graphs, add the following lines to a new cell at the beginning of your notebook. For more information, see Matplotlib's full image tutorial.import matplotlib.pyplot as plt
% matplotlib inline
Q: My IDE shows a Pod in a Bad State error. What causes this error, and how can I fix it?
A: The Pod in a Bad State message is returned when various errors occur:
- Bug
- Missing dependency
- Incorrect configuration
- Lack of memory resources
The IDE might repeatedly try to start and then crash, devolving into a sort of Groundhog Day death loop. We usually need to fix something on our end. Email immunology-support@alleninstitute.org. Include the information listed in the previous question.
Q: How can I execute a shell script within a /private folder?
A: As a workaround, you can copy the shell script to a location within /home/workspace, and then execute the script.
General Questions
Q: Why were the previous-gen HISE IDEs replaced?
A: The previous-gen IDEs were based on the deprecated Google Vertex AI platform. Google has ended support for Vertex AI managed notebooks. Existing instances may continue to function, but patches and updates will not be available. All HISE IDEs have now been migrated to the NextGen IDE platform.
Q: What advantages do the NextGen IDEs offer?
A: NextGen IDEs offer a significantly better user experience compared with the previous-gen IDEs:
- Faster startup
- Billing support
- GitHub integration
- Improved Jupyter notebook UI
- Conda packages composed by key scientists
Future plans include greater disk bandwidth and improved management of idle instances.
Q: How can I report a problem with my IDE and get help from the Immunology Software Dev team?
A: Email your request to immunology-support@alleninstitute.org. Don't worry–this is a monitored inbox, not a cosmic vacuum. A developer will reply during normal business hours. Include the following information in your message:
- IDE name. Provide the name you chose for your IDE.
- Account and project name. Include your account name (such as "Original collective longitudinal cohort study") and project name (such as "FH1 MM Treatment Analysis"). For help finding this information, see Work with Your Accounts and Projects (Tutorial).
- Name of your scientific project. Tell us which research project you're involved with, such as "Health and Age study" or "RA study."
- Steps to reproduce the issue. Describe the context of the crash. What were you doing when it happened? Is the problem constant or intermittent? Have you made any recent changes to the IDE?
- Screenshots or code blocks. Include the error message, error logs, or other additional information to help your support engineer diagnose and troubleshoot the issue.
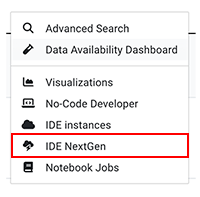
Q: How do I create a NextGen IDE?
A: From the top navigation menu in HISE, click RESEARCH, and then choose IDE NextGen. For detailed instructions, see Create Your First NextGen IDE (Tutorial).
Q: Which folders persist from session to session?
A: See the summary table in Explore NextGen IDE Folders.
Q: Where is the root disk?
A: The root disk is part of the VM instance that's built when you create a NextGen IDE. Because the root disk contains the operating system files, it's sometimes called the "boot disk." When you stop and restart your instance, the root disk persists.
The root disk has a hierarchical folder structure that includes the read-only root directory (/). It also contains writeable paths like /var and /home. The root disk is different from the data disk, /home/jupyter, where you save your files and data.
By default, the size of the root disk is 10 GB. When the disk is full, an HTTP 524 error is returned. For details, see Explore NextGen IDE Folders.
![]()
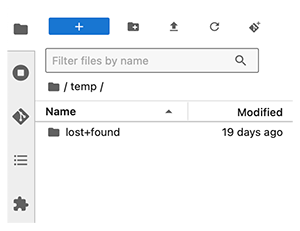
Q: Where is the temporary/scratch disk?
A: The scratch space is your /temp folder. It's used to cache data in active use, process temporary files, and store intermediate computational results. For example, the folder might be used to store data after a system check or repair operation. Files in the /temp folder are persistent. For details, see Explore NextGen IDE Folders.
Q: What is a Conda environment?
A: Conda is a package manager used to bundle source code, tools, dependencies, and instructions for deploying these resources efficiently. When an IDE is created or restarted, Conda quickly spins up an environment based on the selected data modality. For details, see Manage Packages in HISE NextGen IDEs. Conda packages are maintained in a GitHub repository to ensure reproducibility of the steps in the analysis as well as the identical development environment used to obtain a given set of results.
Q: Why is Conda-based installation recommended?
A: Conda creates a snapshot of the tools and resources necessary to re-create scientific or data analytic results. It captures not only the results themselves, but also the environment in which the steps were taken or computations made. The Conda environment includes the code needed to reproduce the exact version of every package and dependency in use when the researchers reached their conclusions. The original researchers or unaffiliated investigators can then attempt to reproduce those results simply by launching the same Conda environment. Conda packages are maintained in a GitHub repository to ensure reproducibility of the steps in the analysis as well as the identical development environment used to obtain a given set of results.
Q: Can I resize an instance?
A: Yes.
Q: If a file goes missing, is it gone forever?
A: No. It's recoverable for about a week. Contact immunology-support@alleninstitute.org for assistance.
Q: When I do a readFiles() or cacheFiles() SDK call, the downloaded files are saved to what location?
A: The files go to the /input folder (/home/workspace/input) in a nested format, as in the following example. For details, see Explore NextGen IDE Folders.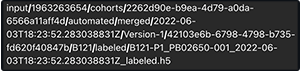
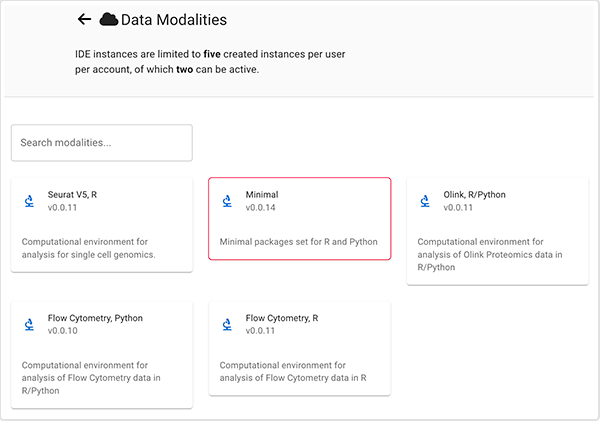
Q: Are R and Python available within a single environment?
A: Yes. To choose R and Python within a single environment, choose "Minimal modality" when you create your IDE.
 Related Resources
Related Resources
Best Practices for NextGen IDE Users
Manage Packages in HISE NextGen IDEs