Understand Automated Pipelines
 | Abbreviations Key |
| API | application program interface |
| HISE | Human Immune System Explorer |
| IDE | integrated development environment |
| LIMS | laboratory information management system |
| QC | quality control |
At a Glance
Pipelines typically start with an automatically triggered ingest process. The exact number of additional steps and the specific processing details vary, depending on the assay type. However, pipelines generally share certain characteristics and follow the same overall workflow.
Pipeline Characteristics
The following table describes the qualities most automated pipelines have in common:
| Characteristic | Description |
| Validation | The data is checked to make sure it's in the expected format and doesn't contain illegal values. An assay-specific scientific QC check is sometimes also performed. |
| Sample association | Assay data will be associated with the samples they belong to. This allows a researcher to search for this data at a later stage by looking for metadata on the sample or the subject or cohort associated with that sample |
| Data preservation | Raw data is automatically preserved in its original state and archived as it enters the pipeline. As new data is generated, data sets and files are created and moved to designated cloud locations, such as MongoDB or private folders, for temporary or permanent storage, or for analysis. |
| Compartmentalization | If the science related to a particular analysis approach changes, isolation of tasks makes it possible to rerun only a few substeps, rather than rerunning the entire pipeline sequence anew. |
| Visual interpretation | Pipeline results files can be read into an IDE or further interpreted using visualization tools. |
Pipeline Workflow
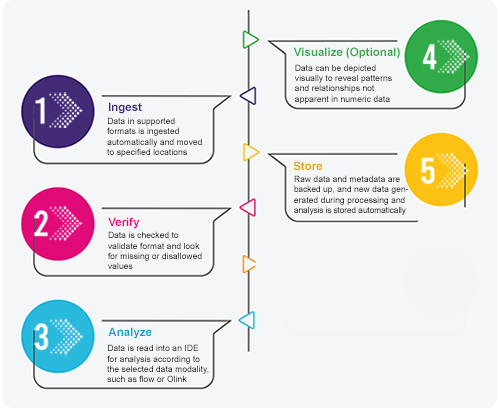
The workflow of an automated pipeline is divided into several steps. We present the steps sequentially here for simplicity, but the workflow isn't strictly linear. Data validation, for example, occurs when the data is ingested, but also when derivative outputs and processed data are returned. Likewise, data storage occurs at multiple points, not just when the transformed data and visualizations are saved.
 Ingest data
Ingest data
In an automated pipeline, polling mechanisms are used to ingest source data automatically. At predetermined intervals or when triggered, these mechanisms interrogate external sources for new data. External sources include, for example, assay results of collected samples in LIMS, partner-managed databases, and questionnaire software with request API integration. Ingest receipts show which data was ingested. These receipts report any errors and flag any missing information.
 Verify
Verify
The accuracy, completeness, and format of source data, intermediate outputs, and processed data must be validated. HISE supports data formats ranging from binary to text to database query syntax. To prevent propagation of errors to subsequent steps in the pipeline, the verification process validates data types and structures, acceptable ranges, cross-field consistency, and other data quality criteria.
 Analyze
Analyze
Lab results and other data can be read into an IDE for analysis according to the selected data modality. At this stage, scientists or analysts use sophisticated computational methods to explore the data. For example, they might identify biomarkers, analyze differential protein expression, or create scripted workflows. Sometimes they generate visualizations to investigated selected datasets, as described in the next step.
 Visualize (Optional)
Visualize (Optional)
It often useful for researchers to read their data into an IDE for interpretation using visualization tools like Plotly and Dash. For example, researchers can use either a subjects query or a samples query to save human metadata, or they can use a result files query to save visualizations of the flow cytometry supervised gating pipeline.For details, see Create a Visualization and Use the HISE SDK to Create Visualizations (Tutorial).
 Store
Store
Automated pipelines store data at every point in the process. At ingest, for example, raw data is archived to capture its provenance and facilitate reproducibility. During data transformation, files might be stored in a cache or staging area. Processed data is stored in various locations for analysis. Storing data at multiple points and in various locations promotes transparency, reproducibility, scalability, and fault tolerance throughout the workflow.
 Related Resources
Related Resources
Configure a Pipeline (Tutorial)