Sample collection and processing
Blood draws from adult donors were processed to isolate peripheral blood mononuclear cells (PBMCs) using a Ficoll gradient within 4 hours after draw, based on our previous study of blood processing delay (Savage, et al. (2021)). Samples were resuspended in FBS with 10% dimethyl sulfoxide (DMSO), and were aliquoted and frozen for shipping to the Allen Institute for later processing in batches.
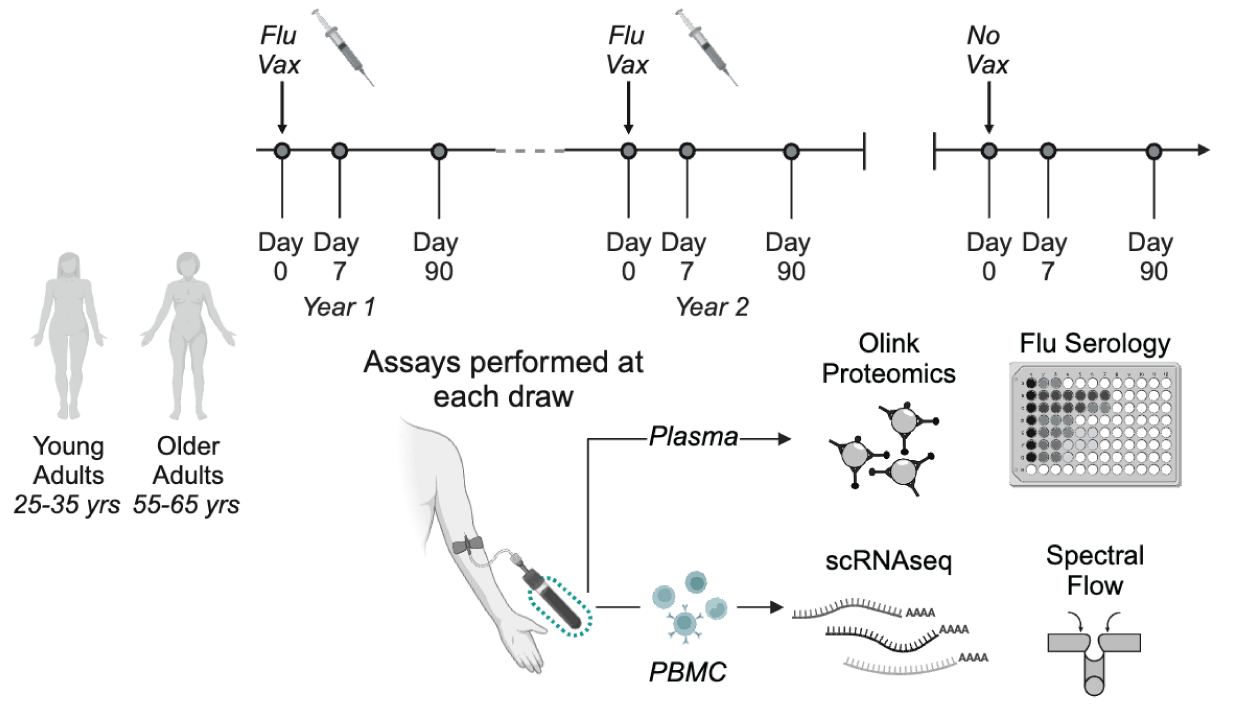
Samples were collected longitudinally to follow changes in peripheral immune state over time. Subjects were administered annual influenza vaccinations (Flu Vax), and samples were collected on the day of vaccination, 7 days post-vaccination, and 90 days post-vaccination. In addition to vaccine time courses, we also collected "No Vax" samples, which were collected with the same timing as flu vaccination sampling, but during periods when no vaccine was administered.
Created with BioRender.com
Flow cytometry and scRNA-seq of batches of samples
Frozen PBMC aliquots were thawed and processed in batches of 12-23 samples, plus a uniform batch control sample, as described in Genge, et al. (2021). After thawing, cells were washed, resuspended, counted, and divided for parallel processing using four panels for high dimensionality spectral flow cytometry (described in Heubeck, et al. (2022)) as well as multiplexed scRNA-seq using 10x Genomics 3' scRNA-seq with Cell Hashing. Pipelines batches contained samples from multiple cohorts collected across projects for the Allen Institute for Immunology. For additional details of our paired experimental pipeline, see our Multiplexed scRNA-seq Pipeline documentation and Genge, et al. (2022).
HCMV serology
Human Cytomegalovirus (HCMV) serology tests were performed at the University of Washington’s Clinical Virology Laboratory in the Department of Laboratory Medicine (https://depts.washington.edu/uwviro/). Plasma or serum samples (200 µL) collected in tandem with blood samples were analyzed using the FDA-approved LIAISON® CMV IgG Assay to detect CMV IgG class antibodies. Results consisted of a CMV Ab Screen Index Value ranging from <0.20 to >10.00, as well as a ‘Positive’ or ‘Negative’ call for anti-CMV IgG detection.
Flu serology
We utilized the Meso Scale Discovery (MSD) platform to asses influenza serology from plasma samples. Total influenza-specific antibody was measured using the MSD Prototype influenza 7-plex Serology Assay to measure antibodies specific for flu vaccine hemagglutinin (HA) antigens: A/Brisbane, A/Hong Kong, A/Michigan, A/Victoria, B/Colorado, B/Phuket, and B/Washington. HA reference standards were used to enable quantification of test samples in units of AU/mL. Neutralizing antibodies were measured using the MSD 96-well hemagglutination inhibition (HAI) 9-plex Assay to measure responses to A/Brisbane, A/Cambodia, A/Guangdong, A/HongKong, A/Kansas, A/Shanghai, A/Wisconsin, B/Phuket, and B/Washington influenza lineages. Results are reported as Percent Inhibition relative to diluent only controls.
Plasma Proteomics
Plasma proteomics assays were performed using the Olink Explore 1536 platform. This platform utilizes paired antibody proximity extension (PEA) coupled to next generation sequencing (NGS) to measure relative expression of 1472 protein targets. We placed longitudinal samples from the same participant on the same plate, and sample position was randomized across plates to avoid positional affects confounding analysis of age and sex. To enable comparisons between plates, we utilized a set of the same plasma bridging controls (12-40) in each of 6 batches.
References
Genge PC, Roll CR, Heubeck AT, Swanson E, Kondza N, Lord C, et al. Optimized workflow for human PBMC multiomic immunosurveillance studies. STAR Protoc. 2021;2: 100900.
doi:10.1016/j.xpro.2021.100900
Heubeck A, Savage A, Henderson K, Roll C, Hernandez V, Torgerson T, et al. Cross-platform immunophenotyping of human peripheral blood mononuclear cells with four high-dimensional flow cytometry panels. Cytometry A. 2023;103: 500–517.
doi:10.1002/cyto.a.24715
Savage AK, Gutschow MV, Chiang T, Henderson K, Green R, Chaudhari M, et al. Multimodal analysis for human ex vivo studies shows extensive molecular changes from delays in blood processing. iScience. 2021;24: 102404.
doi:10.1016/j.isci.2021.102404