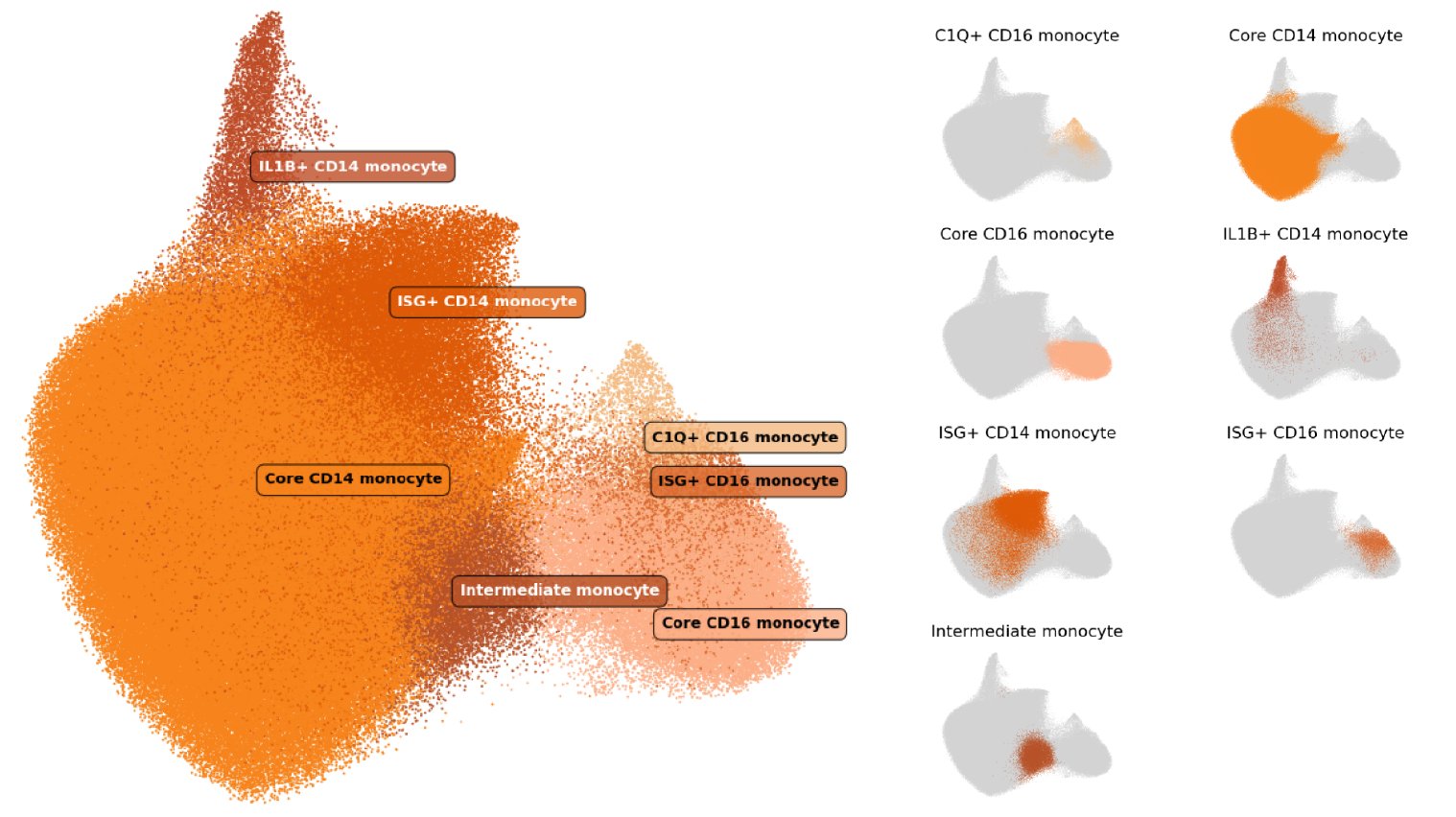
In our cell type atlas, we define Monocyte cell types at 3 levels of resolution, from one Level 1 type (broadest) to 7 Level 3 types (highest resolution) based on marker gene expression and cell types previously described in literature. Descriptions of the marker expression criteria are provided below.
The hierarchical relationships between types, the usage of labels at each level of our cell type hierarchy, and the number and proportion of cells relative to parent populations and the full dataset (All) are shown here:
| Cell Type | Level(s) | N cells | % of Parent | % of All |
|---|---|---|---|---|
| Monocyte | 1 | 327,919 | NA | 18.00% |
| ├ CD14 monocyte | 2 | 269,328 | 82.13% | 14.78% |
| ┆ ├ Core CD14 monocyte | 3 | 217,576 | 80.79% | 11.94% |
| ┆ ├ IL1B+ CD14 monocyte | 3 | 9,046 | 3.36% | 0.50% |
| ┆ └ ISG+ CD14 monocyte | 3 | 42,706 | 15.86% | 2.34% |
| ├ CD16 monocyte | 2 | 45,,920 | 14.00% | 2.52% |
| ┆ ├ Core CD16 monocyte | 3 | 36,364 | 79.19% | 2.00% |
| ┆ ├ C1Q+ CD16 monocyte | 3 | 3,891 | 8.47% | 0.21% |
| ┆ └ ISG+ CD16 monocyte | 3 | 5,665 | 12.34% | 0.31% |
| └ Intermediate monocyte | 2, 3 | 12,671 | 3.86% | 0.70% |
Cell type definitions
Level 1
In this reference we have one Level 1 label:
- Monocyte: The Monocyte class was identified based on high expression of the genes encoding ficolin 1 (FCN1) and cathepsin S (CTSS). Because these genes are also expressed in some dendritic cell (DC) types, we can distinguish monocytes from DCs by low expression of the cDC2 marker CD1c (CD1C).
Level 2
At Level 2, we subdivided Monocytes into 3 major subclasses that are identified and most studied in PBMCs by surface protein expression of CD14, CD16, HLA-D, and CCR2 (Schmidl, et al. 2014; Villani, et al. 2017; Kapellos, et al. 2019).
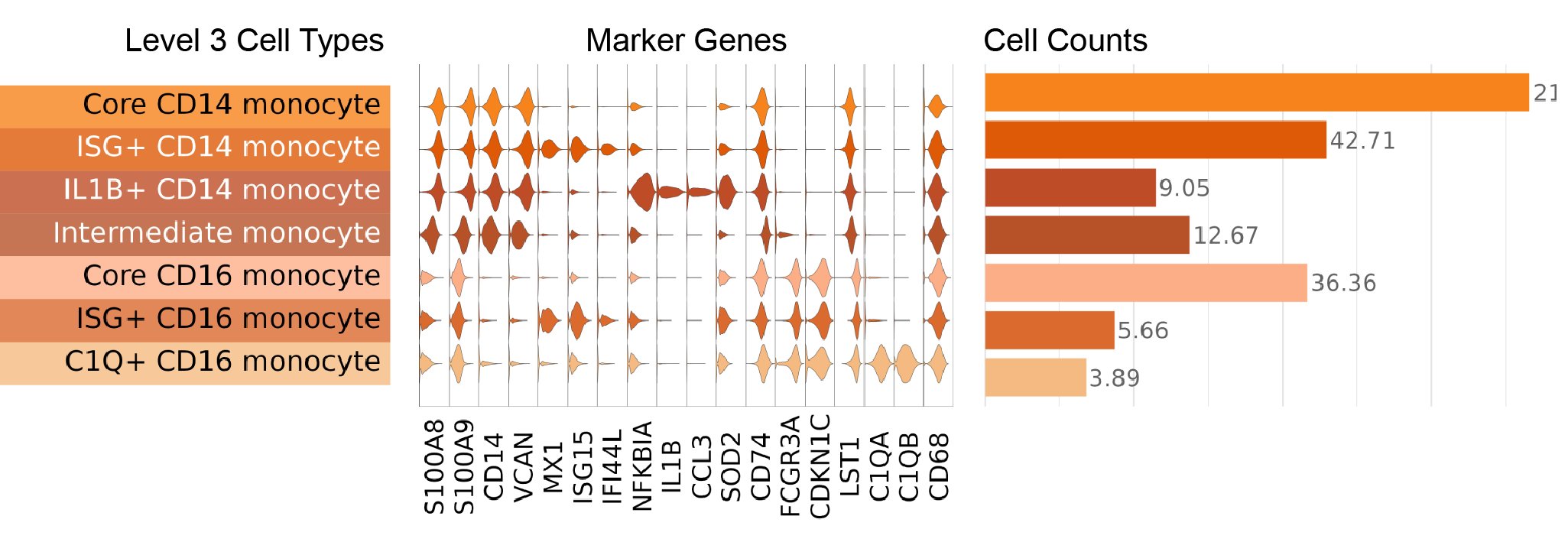
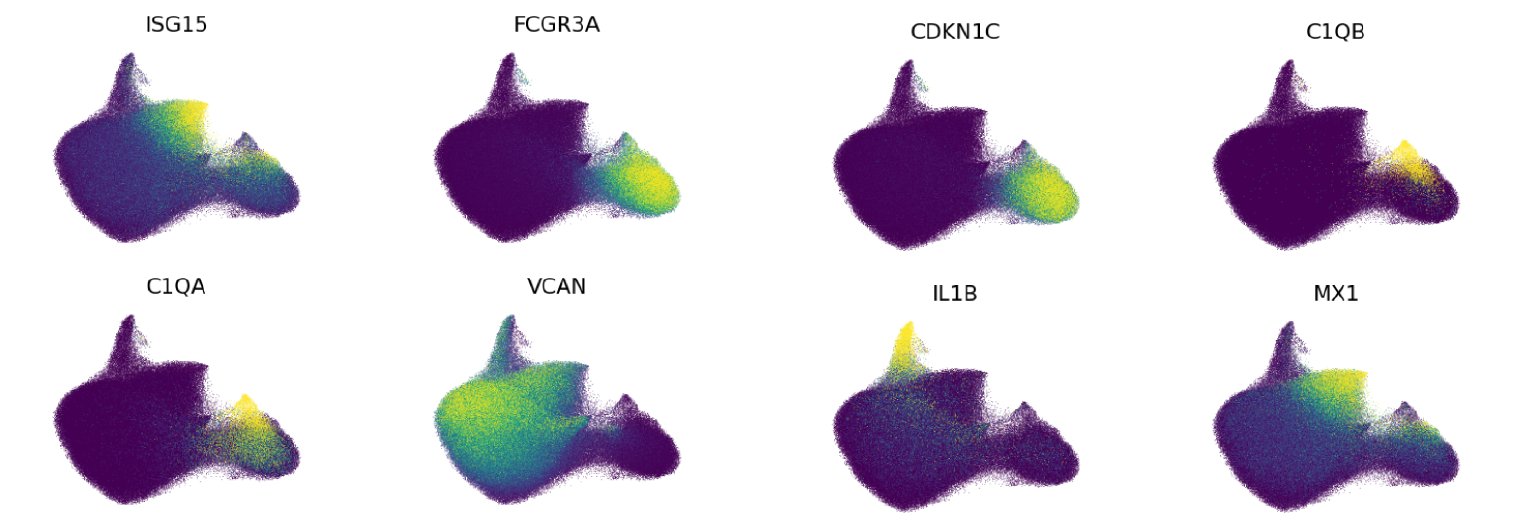
- CD14 monocyte: CD14 monocyte is the most abundant monocyte subset. CD14 monocytes are distinguished from the other Level 2 monocyte cell types by higher expression of the CD14 gene, versican (VCAN), and S100 calcium binding proteins A8 and A9 (S100A8 and S100A9). CD14 monocytes had lower expression of the Fc gamma receptor IIIa gene (FCGR3A) than CD16 monocytes, and lower major histocompatibility complex, class II, DR alpha (HLA-DRA) expression than Intermediate monocytes.
- CD16 monocyte: CD16 monocytes are distinguished by high expression of the gene encoding the CD16 surface marker (FCGR3A), as well as cyclin dependent kinase inhibitor 1C (CDKN1C), and leukocyte specific transcript 1 (LST1).
- Intermediate monocyte: Intermediate monocytes are identified based on their double-positive surface expression of CD14 and CD16. In addition to coexpression of the corresponding CD14 and FCGR3A genes, Intermediate monocytes have higher expression of major histocompatibility complex, class II genes including HLA-DAP1, HLA-DOA, HLA-DRA, and CD74 compared to CD14 and CD16 monocyte types.
Level 3
At Level 3 of our annotations, we further divided the CD14 and CD16 monocytes based on expression of specific gene programs. Intermediate monocytes were not subdivided further.
CD14 monocytes were divided into 3 subpopulations:
- IL1B+ CD14 monocyte: IL1B+ CD14 monocytes are a relatively rare subset of CD14 monocytes that have been recently shown to expand in the blood of checkpoint blockade-induced inflammatory arthritis patients (Zhou, et al. 2024). In addition to possessing CD14 monocyte transcriptome signatures, IL1B+ CD14 monocytes express high levels of the interleukin 1 beta gene (IL1B), the C-C motif chemokine ligand 3 gene (CCL3), and the C-X-C motif chemokine ligand 8 gene (CXCL8).
- ISG+ CD14 monocyte: Similar to many other key immune subsets in our atlas, a subset of CD14 monocytes display a strong interferon-stimulated gene (ISG) transcriptome signature. These cells express high levels of ISGs including MX dynamin like GTPase 1 (MX1), interferon induced protein 44 like (IFI44L), and interferon alpha inducible protein 6 (IFI6). A recent report also identified this population in human PBMCs and shows it is a stable monocyte subset that not only exists in healthy subjects, but also in cancer patients (Affandi, et al. 2021).
- Core CD14 monocyte: The remaining CD14 monocytes that did not express the distinct transcriptome signatures for IL1B+ or ISG+ CD14 monocytes were assigned the Core CD14 monocyte label.
CD16 monocytes were divided into 3 subpopulations:
- C1Q+ CD16 monocyte: C1Q+ CD16 monocytes are a cell type that has been recently reported in subjects with Behçet’s disease, and may mediate hyperinflammation in this setting (Zheng, et al. 2022). However, we also identify these cells as a small subset of CD16 monocytes in our healthy cohorts across age. These monocytes are distinguished by high expression of the complement C1q A chain and B chain genes (C1QA and C1QB, respectively)
- ISG+ CD16 monocyte: As described for CD14 monocytes above, we identify an analogous ISG+ cell type among CD16 monocytes that express ISGs including MX1, IFI44L, and IFI6.
- Core CD16 monocyte: The remaining CD16 monocytes that did not express the distinct transcriptomic signatures for C1Q+ or ISG+ CD16 monocytes were assigned the Core CD16 monocyte label.
Marker visualizations
Key markers used to define NK and ILC cell types are shown in this figure, where we display a violin plot for each marker gene and each Level 3 cell type:
Expression of key marker genes shown on our UMAP projection:
References
Affandi AJ, Olesek K, Grabowska J, Nijen Twilhaar MK, Rodríguez E, Saris A, et al. CD169 defines activated CD14+ monocytes with enhanced CD8+ T cell activation capacity. Front Immunol. 2021;12: 697840.
doi:10.3389/fimmu.2021.697840
Kapellos TS, Bonaguro L, Gemünd I, Reusch N, Saglam A, Hinkley ER, et al. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol. 2019;10: 2035.
doi:10.3389/fimmu.2019.02035
Schmidl C, Renner K, Peter K, Eder R, Lassmann T, Balwierz PJ, et al. Transcription and enhancer profiling in human monocyte subsets. Blood. 2014;123: e90-9.
doi:10.1182/blood-2013-02-484188
Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356.
doi:10.1126/science.aah4573
Zheng W, Wang X, Liu J, Yu X, Li L, Wang H, et al. Single-cell analyses highlight the proinflammatory contribution of C1q-high monocytes to Behçet’s disease. Proc Natl Acad Sci U S A. 2022;119: e2204289119.
doi:10.1073/pnas.2204289119
Zhou Z, Zhou X, Jiang X, Yang B, Lu X, Fei Y, et al. Single-cell profiling identifies IL1Bhi macrophages associated with inflammation in PD-1 inhibitor-induced inflammatory arthritis. Nat Commun. 2024;15: 2107.
doi:10.1038/s41467-024-46195-x