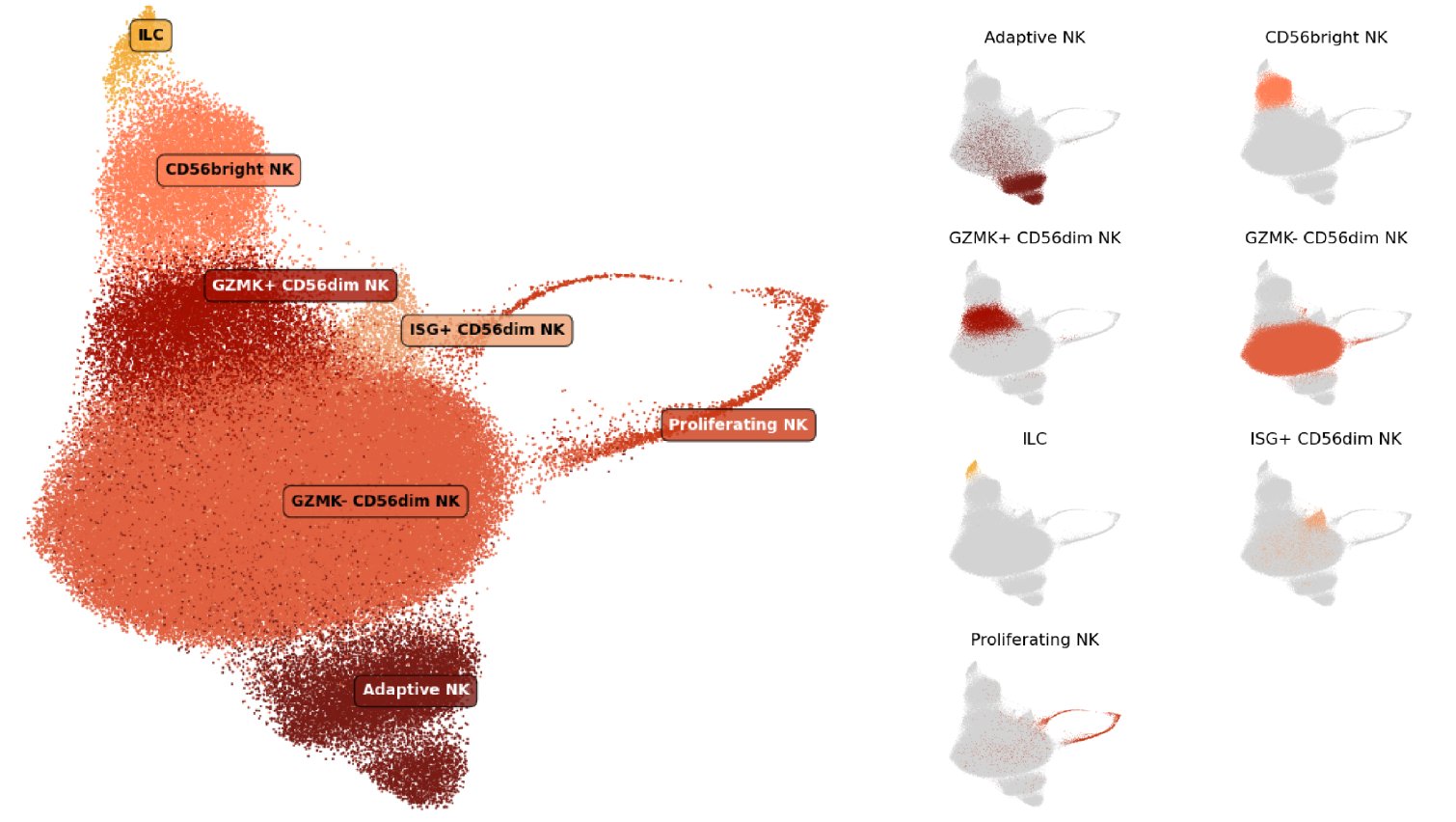
In our cell type atlas, we define Natural Killer (NK) cell and innate lymphoid cell (ILC) cell types at 3 levels of resolution, from two Level 1 types (broadest) to 7 Level 3 types (high resolution) based on marker gene expression and cell types previously described in literature. Descriptions of the marker expression criteria are provided below.
The hierarchical relationships between types, the usage of labels at each level of our cell type hierarchy, and the number and proportion of cells relative to parent populations and the full dataset (All) are shown here:
| Cell Type | Level(s) | N cells | % of Parent | % of All |
|---|---|---|---|---|
| ILC | 1, 2, 3 | 844 | NA | 0.05% |
| NK cell | 1 | 147,761 | NA | 8.11% |
| ├ CD56bright NK cell | 2, 3 | 11,055 | 7.48% | 0.61% |
| ├ CD56dim NK cell | 2 | 133,881 | 90.61% | 7.35% |
| ┆ ├ GZMK- CD56dim | 3 | 102,908 | 76.87% | 5.65% |
| ┆ ├ GZMK+ CD56dim | 3 | 12,752 | 9.53% | 0.70% |
| ┆ ├ Adaptive NK cell | 3 | 14,735 | 11.01% | 0.81% |
| ┆ └ ISG+ CD56dim NK cell | 3 | 3,486 | 2.60% | 0.19% |
| └ Proliferating NK cell | 2, 3 | 2,825 | 1.91% | 0.12% |
Cell type definitions
Level 1
NK cells are classically divided into two major subsets based on cell surface detection of CD56 and CD16 in flow cytometry: CD56brightCD16low and CD56dimCD16hi. In this reference we have two Level 1 labels:
- NK cell: For use at the broadest level of our Immune Health Atlas, Level 1, these two classes are combined to define NK cell as a label encapsulating both classical subclasses.
- ILC: We identify a population at all levels of our type hierarchy for ILCs. While further division of subpopulations of ILCs is possible (Spits and Cupedo, 2012), the low abundance of circulating ILCs available in our PBMC dataset prevented further segregation of subclasses.
Level 2
We retain the two classically defined groups, above, in our Level 2 annotations, as well a group of Proliferating NK cells:
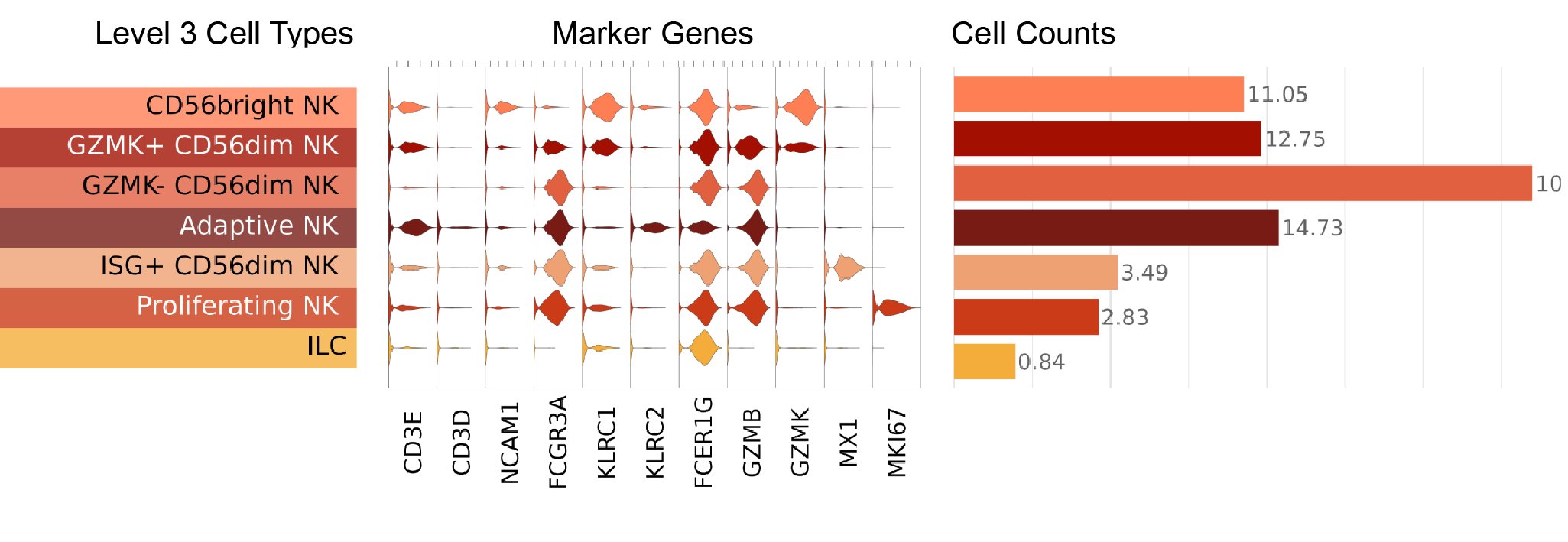
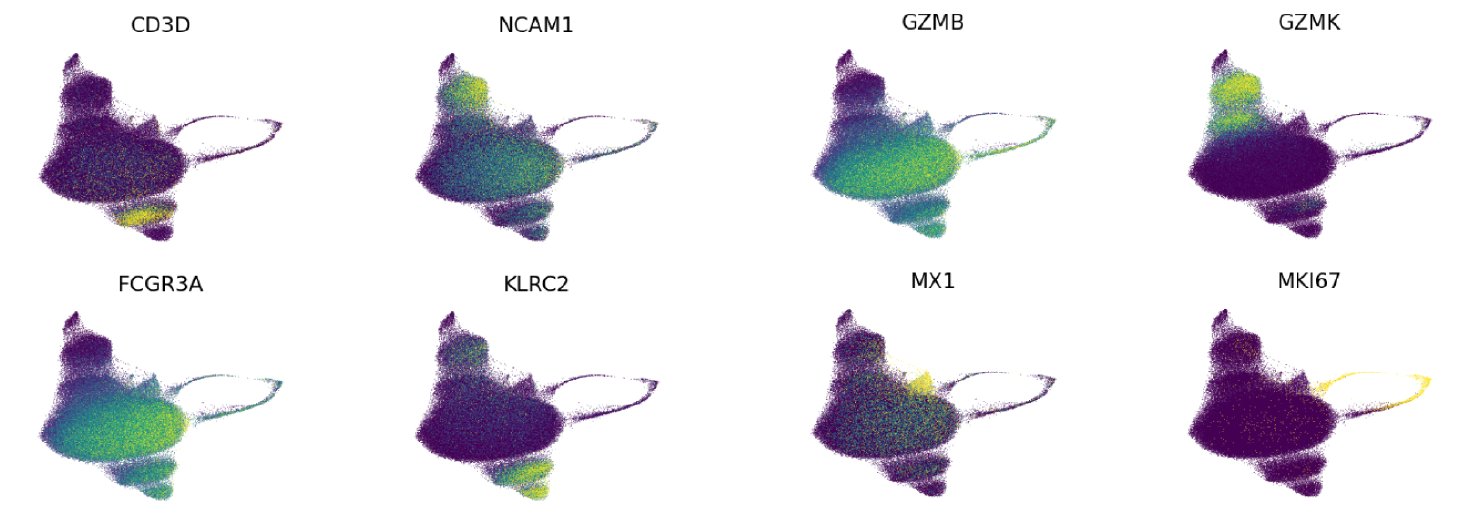
- CD56dim NK cell: CD56dimCD16hi clusters were assigned the CD56dim NK cell label based on low expression of the neural cell adhesion molecule 1 (NCAM1) gene and high expression of the Fc gamma receptor IIIa (FCGR3A) gene, which encode CD56 and CD16, respectively.
- CD56bright NK cell: CD56brightCD16low clusters were assigned the CD56bright NK cell label based on high NCAM1 and low FCGR3A expression.
- Proliferating NK cell: Proliferating NK cells were identified as clusters with high expression of the marker of proliferation Ki-67 (MKI67) gene.
Level 3
At Level 3 of our annotations, we further divided the CD56dim NK cells based on expression of specific gene programs:
- GZMK+ CD56dim NK cell: Cell clusters expressing the granzyme K (GZMK) gene were assigned the label GZMK+ CD56dim NK cell.
- Adaptive NK cell: CD16-positive clusters that also expressed high levels of killer cell lectin like receptor C2 (KLRC2) transcription were assigned the Adaptive NK cell label. Adaptive NK cells have been described as a type that expands in response to CMV infection in mice (Sun, et al. 2009) and humans (Lopèz-Verges, et al. 2011).
- ISG+ CD56dim NK cell: Clusters with high expression of interferon-stimulated genes (ISG), including ISG15 ubiquitin like modifier (ISG15), and MX dynamin like GTPase 1 and 2 (MX1, and MX2, respectively) were assigned the ISG+ CD56dim NK cell label.
- GZMK- CD56dim NK cell: Remaining CD56dim NK cells that did not express the markers above formed a uniform group of cells that were assigned the GZMK- CD56dim NK cell label.
Marker visualizations
Key markers used to define NK and ILC cell types are shown in this figure, where we display a violin plot for each marker gene and each Level 3 cell type:
Expression of key marker genes shown on our UMAP projection:
References
Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2011;108: 14725–14732.
doi:10.1073/pnas.1110900108
Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30: 647–675.
doi:10.1146/annurev-immunol-020711-075053
Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457: 557–561.
doi:10.1038/nature07665